Chapter 8 MA Plots
MA plots are very useful in interpretation of the data as they visualise genes that are highly abundant and most different between two conditions/treatments. These MA plots are automatically labelled with gene names of the outermost genes on the plot. It also identifies PIF and DE genes. In fact, the PIF dataset is used as input to this function as it contains all genes ranked on PIF, as well as p values for filtering DE genes.
Note that this function is a wrapper to the plot_custom_MA() function within DE4Rumi and any additional arguments from plot_custom_MA can also be parsed to the auto_plot_multi_MA function. This allows for significant customisation, although the default settings are likely sufficient. For example, tiff_width= and tiff_height= are valid arguments but leaving these out means their default values are used. In addition, the thresholds for DE and PIF can be customised. See ?plot_custom_MA.
#access PIF data
PIF_dataframes <-
DE_out %>%
purrr::map("PIF") %>%
unlist(recursive = FALSE)
MA_plots_out <-
auto_plot_multi_MA(PIF_list = PIF_dataframes,
list_plot_titles = NA,
export_TIFFs = TRUE,
export_pdf = TRUE,
pdf_height = 140,
pdf_width = 220,
export_dir = "./outputs/MA_plots/")
MA_plots_out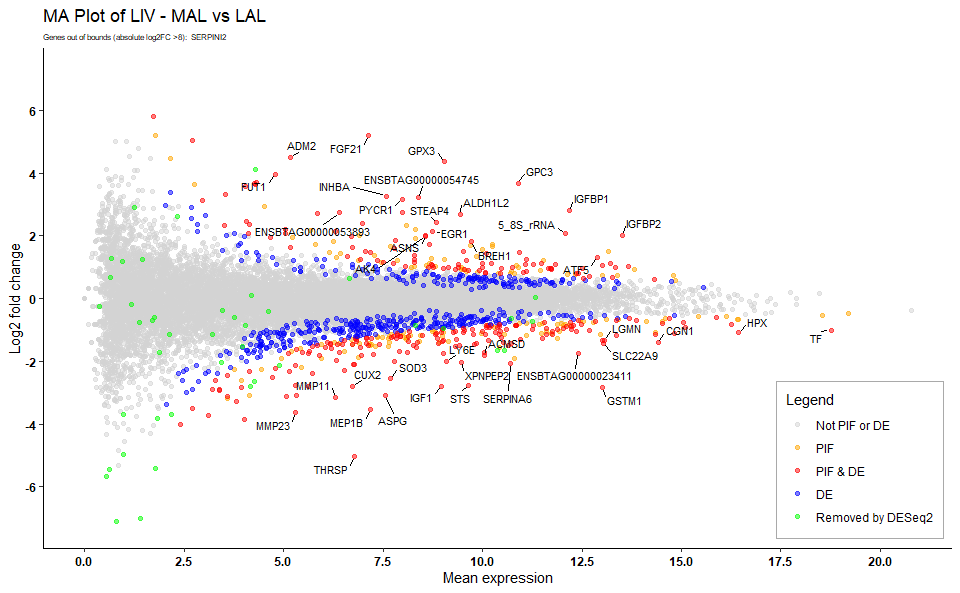
Figure 8.1: Example MA plot.