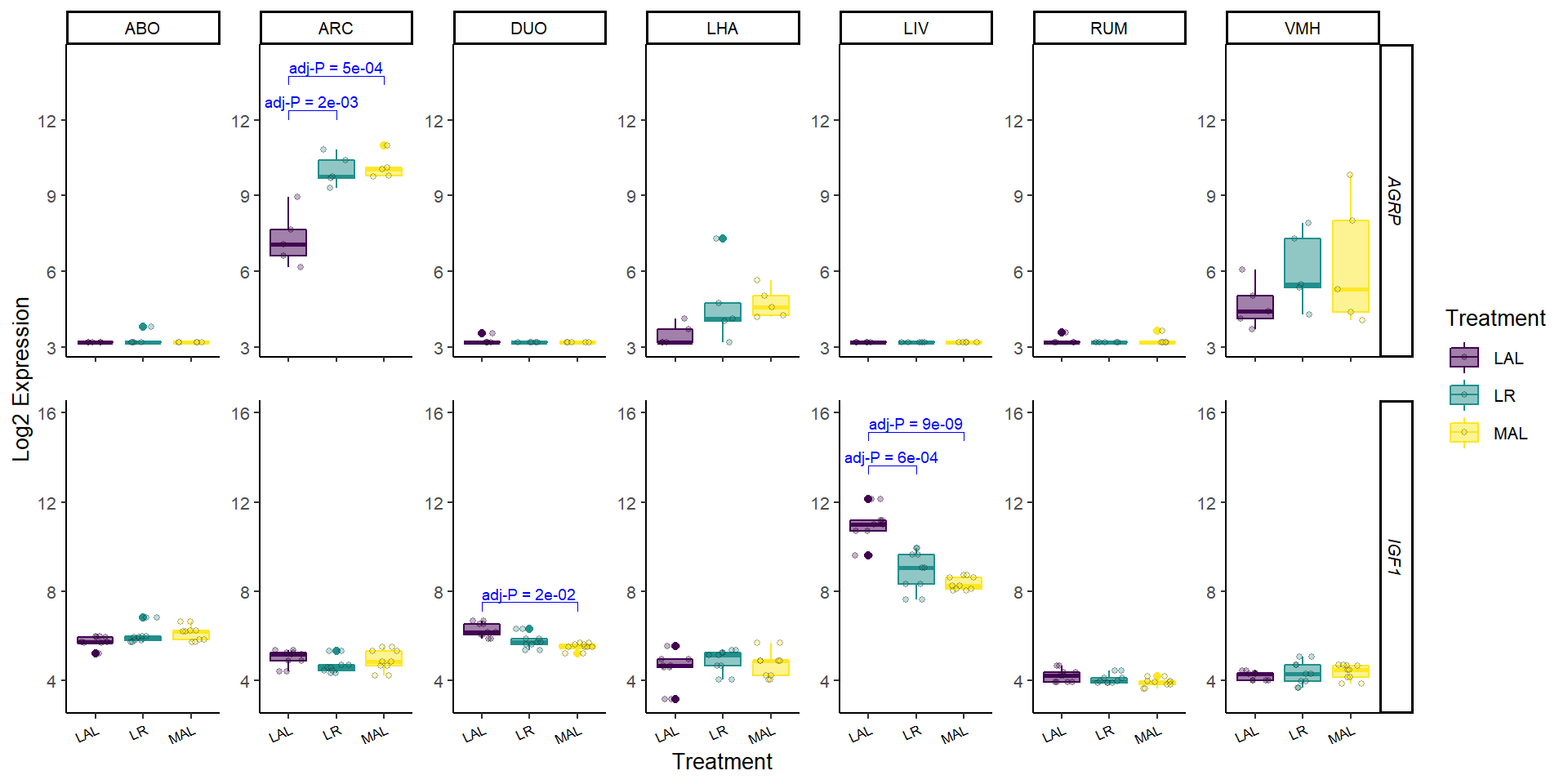
Chapter 12 Plot genes of interest
Sometimes it is useful to view boxplots of the expression of a gene for all individual samples. This function allows genes of interest to be plotted in detail.
Multiple names can be provided. The names of the genes are
groups_for_DE <- seq_data$Tissue_Region %>% unique()
DE4Rumi::plot_genes_of_interest(norm_exp_data = vst_norm_data,
DE_full_out = DE_out,
se_data = seq_data,
gene_annotations = gene_annot,
selected_genes = c("AGRP", "IGF1"),
top_level_colname = Tissue_Region,
sample_colname = sample_names,
results_contrast_factor = Treatment,
top_level_groups = groups_for_DE,
plot_groups_colname = Treatment,
max_genes_per_plot = 3,
x_label = "Treatment",
p_thresh = 0.05)## Joining, by = "sample_names"## Joining, by = "Treatment"
## Joining, by = "Treatment"## Joining, by = c("gene_name", "top_level")
## Joining, by = c("gene_name", "top_level")## $`1`